Papain: A natural protease made by papaya.

Background
Greetings curious readers, if you like biology and organic chemistry i got something interesting to present today. A natural compound with multiple uses and a fully mapped mechanism of action. I will render the protein as good as i can, to make it´s structure and function understandable.
Introduction
Papain is a protein with a protease function meaning that it cleaves peptide bonds at certain positions of a protein.
It has numerous industrial applications in everything from home remedies, cosmetics and toothpaste to industrial usage in meat and biotech sectors as well as the medical industry, which makes it a good enzyme to know about. Toothpastes also add bromelain from Pineapples, another protease. Papain is used with bromelain as meat tenderizers industrially, which is good I guess, if you eat meat. Papain has been studied a lot and the catalytic mechanism has been mapped in detail, which is pretty rare for enzymes in general.
It is linked to many health benefits as well as skin treatments, and the seeds is just as nutritional as the rest, if not more off course! Who doesn't like home remedies ? Check it out in on the link below.
All the molecule representations is made by me, feel free to comment if you need tips or tricks in this area. I like PyMol and mostly use this software, a python program.
Structure
Papain also called papaya proteinase 1, consists of 345 amino acids, making it a medium sized to small protein, but comes with big possibilities to use in biochemistry. It has a signal peptide, residuals 1-18, a pro-peptide, 19-133 and the mature peptide, 134-345.
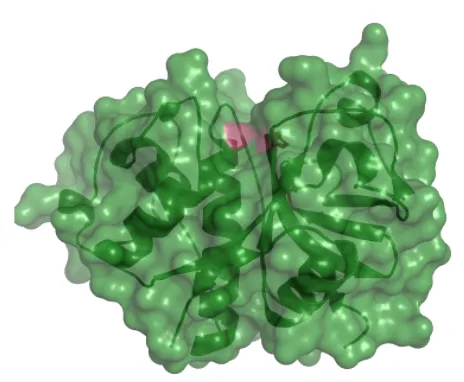
Papain has two subdomains that come together to form the final protein structure looking like the illustration above. It´s active and catalytic site is located between the two domains and is highlighted in red above.
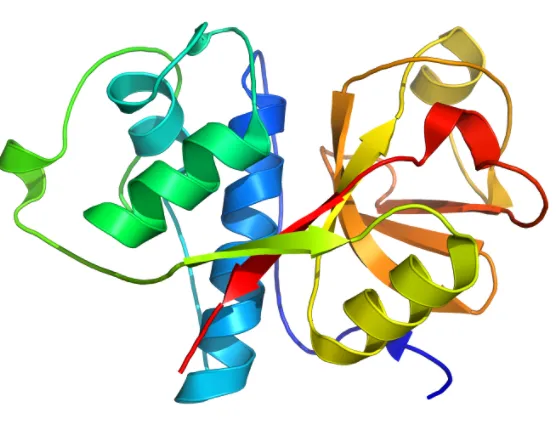
We can see that the two domains is not similar in structure, which is good to notate, since many enzymes with multiple domains can have similar subunits. Hemoglobin for example has 4 identical subunits each with oxygen binding site, but the middle heme binding site is made cooperatively by all four domains. One domain has two large alpha helices seen in blue / cyan, while the other domain have one large alpha helix and a larger beta sheet region, as seen in yellow and orange.
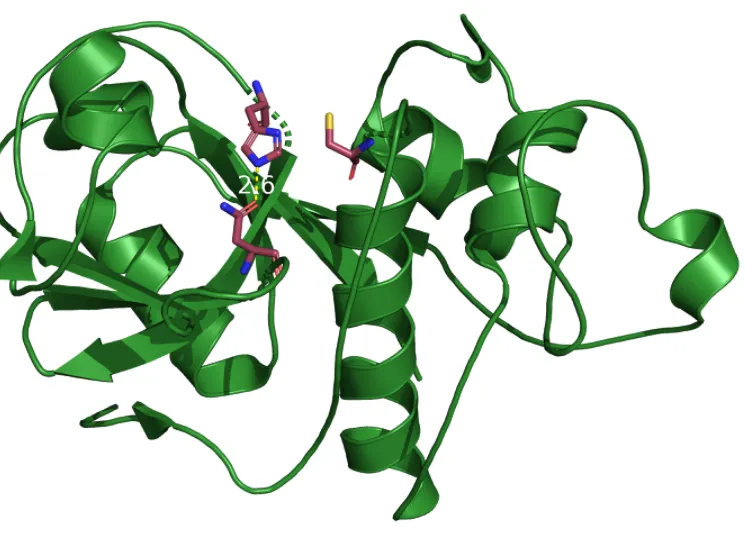
As I mentioned, the catalytic site is well studied and consists of a catalytic tirade which is very common for proteases. Papain is a cysteine protease, meaning that a cysteine residue is involved in performing the peptide cleaving.
Visible above is the three mechanistically important residues. Asparagine 175 orients the ring on the next visible residue, Histidine 159 to allow it to deprotonate the last catalytic residue, cysteine so it can cleave the peptide. The atom color is correct for sulfur, so cysteine is the one to the right of the histidine ring, with a yellow sulfur tip(group).
It is very heat resistant so that perfect for industrial applications, and it actually performs the best between 50-70 degrees!
Mechanism
All the mechanisms is available at https://www.ebi.ac.uk/thornton-srv/m-csa/ which is a merged data base of the old parent databases MACiE( Mechanism, Annotation, Characterization in Enzymes) and CSA(Catalytic Site Atlas) resulting in the database M-CSA( Mechanism and Catalytic Site Atlas) which is a good project, classifying enzymes by actuall known mechanisms. We are at 1000 entries and growing, which is great for the industry of biochemistry!
In step 1, the sulfur on the cysteine is a nucleophile, meaning it like positive groups(nucleus) and will attacks the carbonyl coal on the backbone of the target peptide, which has a positive dipole moment. This results in a nucleophilic addition to the target peptide.
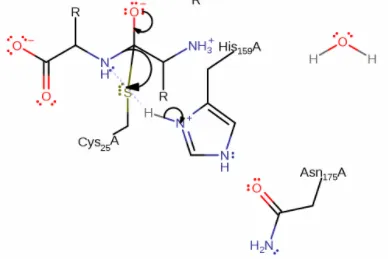
An electron located on the double bond to the oxygen moves to the oxygen generating an oxyanion.
Next step 2 is initiated by the oxyanion initiates an elimination reaction that will cleave the peptide. As you can see above the sulfur steals the electron bound from the carbonyl coal while by help of the oxyanion which will re-generate the double bound to the same carbonyl coal. The deprotonation of cysteine, is essential generating a stronger nucleophile.
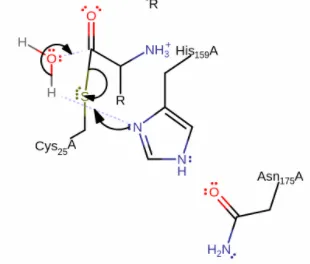
In the 3rd and last step, the histidine instead deprotonates a water molecule, and the electron attacks the same carbonyl coal as in step 1 and 2. This initiates a nucleophilic substitution, swaping the sulfur bond for the water group, regenerating the cysteine to its original state, and leaving an de-protonated carboxylic-acid terminal on the leaving peptide fragments. The peptide is now cleaved and the cysteine back to its original state, ready for additional cleaving of peptides.
This is how we believe that the full procedure of cutting a peptide by Papain at a proteins backbone. All amino acids have the same backbone atoms, N-C-C(nitrogen & carbon) and links to another amino acids and repeats, N-C-C-N-C-C-N-C-C and so on. Papain won´t cut just anywhere and is known to be specific to a hydrophobic region(water hating) residual like Leucine(nonpolar), which is logical since the part that is to be cut must be placed between the two terminals. It also have affinity towards a few specific amino acids like that should follow the hydrophobic one, Arginine and Lysine, and it won´t cut at all if a valine is on any of these three positions. In short it cuts at the three amino acids motif: Hydrophobic-AA/Arg/Lys.
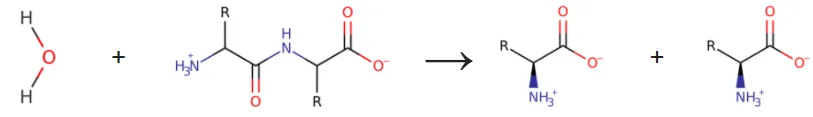
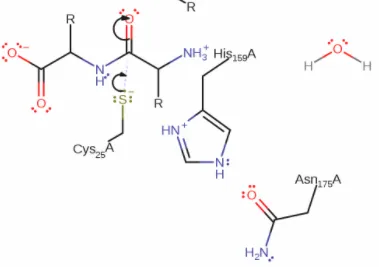
The last picture, above, is a simplification of the full reaction, showing only products and reactants, no mechanisms or steps. Here it is easy to see a peptide(protein) backbone being split in two.
References:
Thanks for reading and supporting my work, it is much appreciated.
