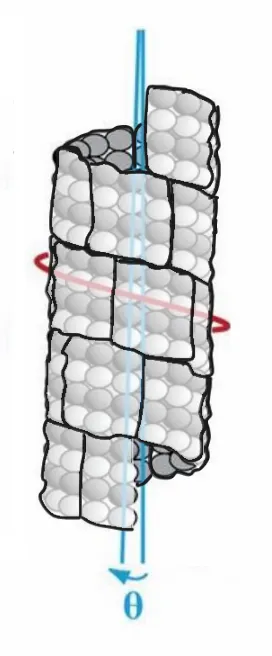
Microtubules are 24 nm in diameter, with a circumference of 75 nm, each tubulin filament occupying 5.8 nm, and with a longitudinal periodicity that of the length of tubulin dimers, 8 nm. The 3 x 2 tubulin lattice has a width of the surface width each tubulin takes up within the microtubule, 5.8 nm x 3 = 17.4 nm, and a height of two tubulin dimers, 8 nm x 2 = 16 nm, approximating a square.

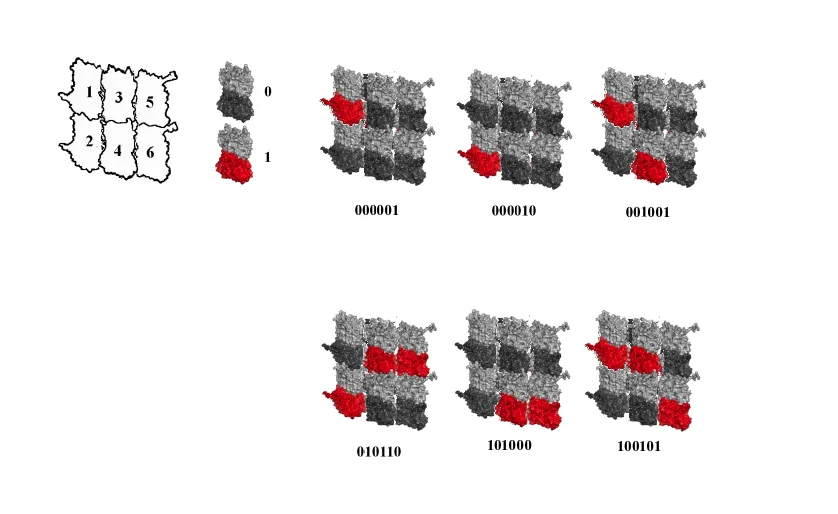
In its activated state, CAMKII is 20 nm, and binds to the 3 x 2 lattice field, writing 6 bits of information, a microtubule byte, with 26 (64) possible states.
The image below shows 6 out of 64 possible phosphorylation states in a 3 x 2 lattice field.
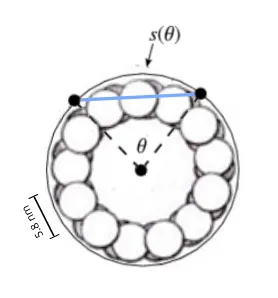
The width of the 3 x 2 lattice field is curved, and the width adjusted for curvature, if CAMKII binds as if binding to a flat surface, is calculated from 2sinθ/2 where θ is the curved width 17.4 nm / microtubule radius of 12 nm, 1.32627 radians, and multiplied with the radius 12 nm for the adjusted width 15.91524 nm, close to the 16 nm height of the 3 x 2 lattice field.
The 3 x 2 lattice field that CAMKII "sees" is 15.9 nm * 16 nm, almost a perfect square.

The 16 x 16 nm square lattice field below a CAMKII with a diameter of 20 nm.

To CAMKII, the outer tubulin columns appear compressed relative to the middle column, each roughly 5.05 nm wide instead of 5.8 nm, from how CAMKII "sees" a 15.9 nm flat projection of the 17.4 nm wide tubulin memory block (r2sinθ/2).
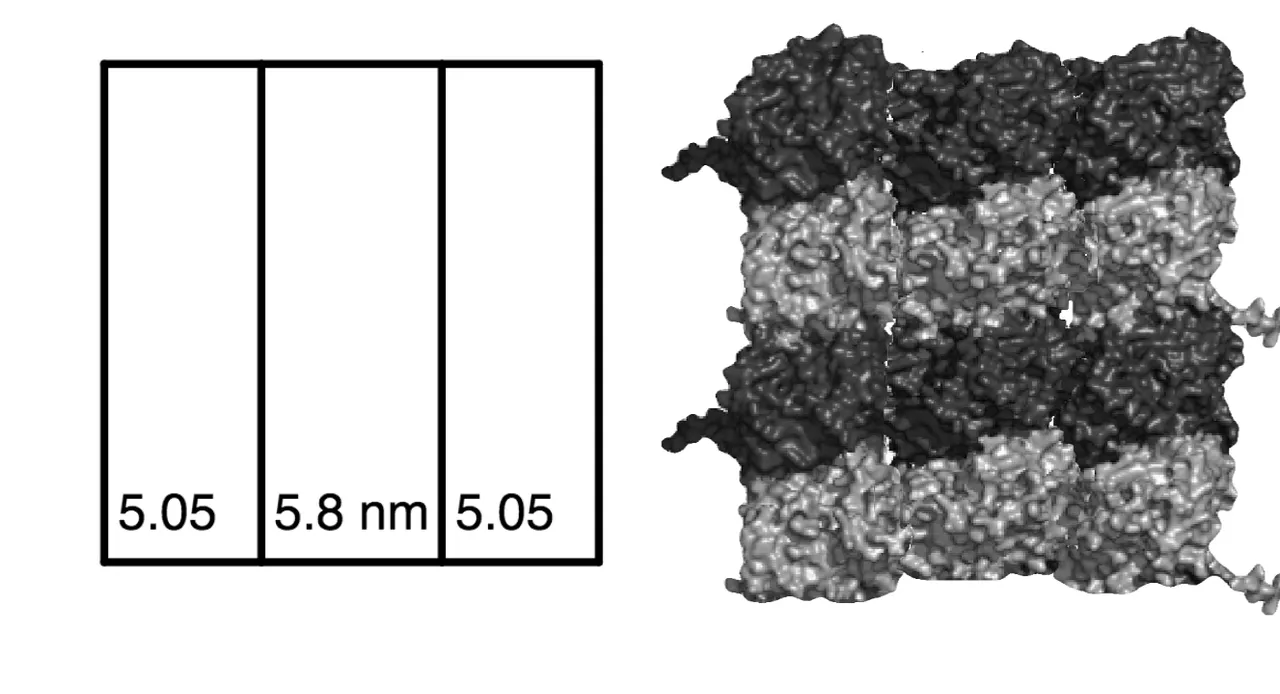
The 6-tubulin "bytes" fit perfectly on a B-lattice. The addressing system could perhaps be built on microtubule-associated proteins that form some type of structure along the MT?